Powerful Deep Learning
EDIT 3/7/2019: The method referenced in the code was later published as a Nature article: https://www.nature.com/articles/nature25988. A possible improvement discussed with the author is complex number support via say via CUTLASS: https://github.com/NVIDIA/cutlass
The most important trend in artificial intelligence this year was the dramatic improvements to accessibility. We seek to unpack this oft parroted “democratization of AI” with a concrete example as well as circle the key developments. The toolset for AI and deep learning includes releases such as Google DeepMind’s Sonnet https://github.com/deepmind/sonnet, Keras incorporated in Tensorflow’s main branch https://github.com/keras-team/keras, and Uber’s Horovod for distributed training https://github.com/uber/horovod to name a few. Taken as a group, these toolkits allow the expression of powerful ideas with simple (re: elegant) execution. In the topic of medical image reconstruction, such examples have arisen (https://medium.com/@LeoKTam/compressed-sensing-and-deep-learning-revisited-414e0161cfe2) and powerfully general medical image reconstruction is possible in few lines of Keras.
Three recent papers on medical image reconstruction “A Deep Cascade of Convolutional Neural Networks for MR Image Reconstruction“ https://arxiv.org/abs/1703.00555, “Deep Generative Adversarial Networks for Compressed Sensing Automates MRI” https://arxiv.org/abs/1706.00051, and “Image reconstruction by domain transform manifold learning” https://arxiv.org/abs/1704.08841 tackle more general problems than previous approaches. The deep cascade solves the problem of high quality reconstruction of arbitrary MRI sampling patterns with rapid speed. Previous iterative approaches would require several minutes while this approach reduced it to 23 ms. Using alternating neural network modules, they controlled data fidelity and denoising terms, refining the reconstruction to an excellent quality in a single processing pipeline. Both the Stanford and Harvard approach abstracted the reconstruction process. The approach from Harvard known as AutoMAP sought to unify medical reconstruction across modalities.
The work from Zhu et. al. in AutoMAP (which we’ll replicate) took a similar approach to “Deep Cascade” by having one portion of the network for data consistency and the other portion for superresolution or refinement of image quality. The neural network architecture which is replicated in Keras in a few lines code first clubs the data transformation problem with several fully connected layers (exploding the parameter count to the hundred of millions) and then followed by a few layers reminiscent of a sparse denoising decoder. The simple expression of data consistency and superresolution solves image reconstruction generally across modality, displacing algorithms such as Kaczmarz algebraic reconstruction, filtered back projection, and iterative conjugate gradients reconstruction. If a criticism is the data-driven requirements, it’s possible to source open data from government funded projects such as the Connectome project https://db.humanconnectome.org, from http://www.mridata.org/, other repositories http://reddit.com/r/dldata, or bootstrap through simulation http://mrirecon.github.io/bart/. As one radiologist related to me at RSNA 2017, often 8-9 reading terminals are crammed into one room. Deep learning based reconstruction technology could unify and disrupt these inefficiencies.
As promised we may describe the AutoMAP architecture in a few lines of Keras code:
Such code combined with an appropriate simulation package generates a MR reconstruction module, figure below.
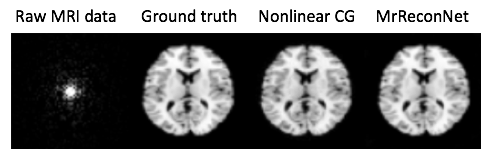
Powerful deep learning tools are now broadly and freely available. In 25 lines of code, we can specify a neural network architecture that supersedes decades of hand-crafted code for image reconstruction across modalities, achieving a “Krizhevsky” of medical image reconstruction. Democratizing AI means powerful tools for all.